Research in the Herr Laboratory
> How our research benefits the public and society:
Members of the Herr Lab work on many different systems, but what unites our microbiological research is the desire to answer the questions: “who is there?” and “what are they doing?”
The end goal of our reseach is to reduce disease prevalence and increase the overall health of humans, plants, animals, and the planet we live on.
> Deeper into the science in our laboratory:
Metagenomics in complex systems

Soil is an incredibly important for environmental health and plant health and productivity. Soil is arguably the most diverse ecosystem on the planet and it holds an amazing array of microbes, both known and unknown, with an amazing array of functional attributes, both known and unknown.
Our lab uses nucleotide sequencing techniques and other methods to characterize both taxonomy and functional diversity of microbes in soil. We work with both bacterial and fungal species and use computational methods to extract genomic information from metagenomic and metatranscriptomic sequencing data.
The goal of this research is to identify microbes which may help reduce disease prevalence in plant systems, increase the ability of a plant to increase uptake of essential nutrients, and allow for a resilient and healthy ecosystem (often threatened by changing climate scenarios).
Understanding Plant-Microbe Interactions
The overall health of plants and their is influenced by microbes, which can be pathogens, mutualists, and commensal organisms. Some microbes may be a pathogen on one plant and a mutualist on another.
How are these associations regulated and what is the mode of interaction between host and microbe?
We use metagenomics, metatranscriptomics, and metaproteomic approaches to address these relationships. Our goal is to understand the mechanisms that regulate plant-associated microbial communities.
Comparative Genomics and Metagenomics
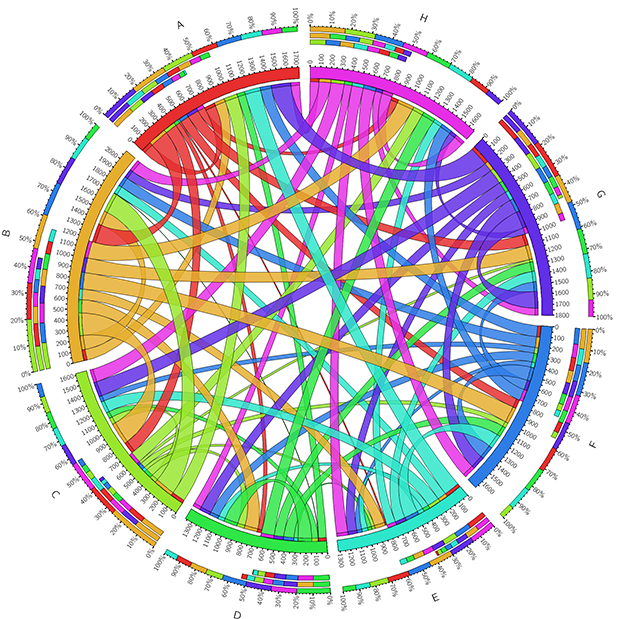
Using genome sequencing techniques, we are inspecting the taxonomic and functional diversity from single sequenced genomes and genomes assembled from mixed metagenomic reads.
Using these genomes, we’re investigating the role of the pan-genomics of many types of organisms, such as fungi, bacteria, and viruses. By studying both the core and accessory genes in these genomes we hope to elucidate both symbiotic and pathogenic relationships with hosts. Candidate genes for these roles include effectors and other small secreted proteins.
Understanding the taxonomic and functional diversity of mycorrhizal fungi
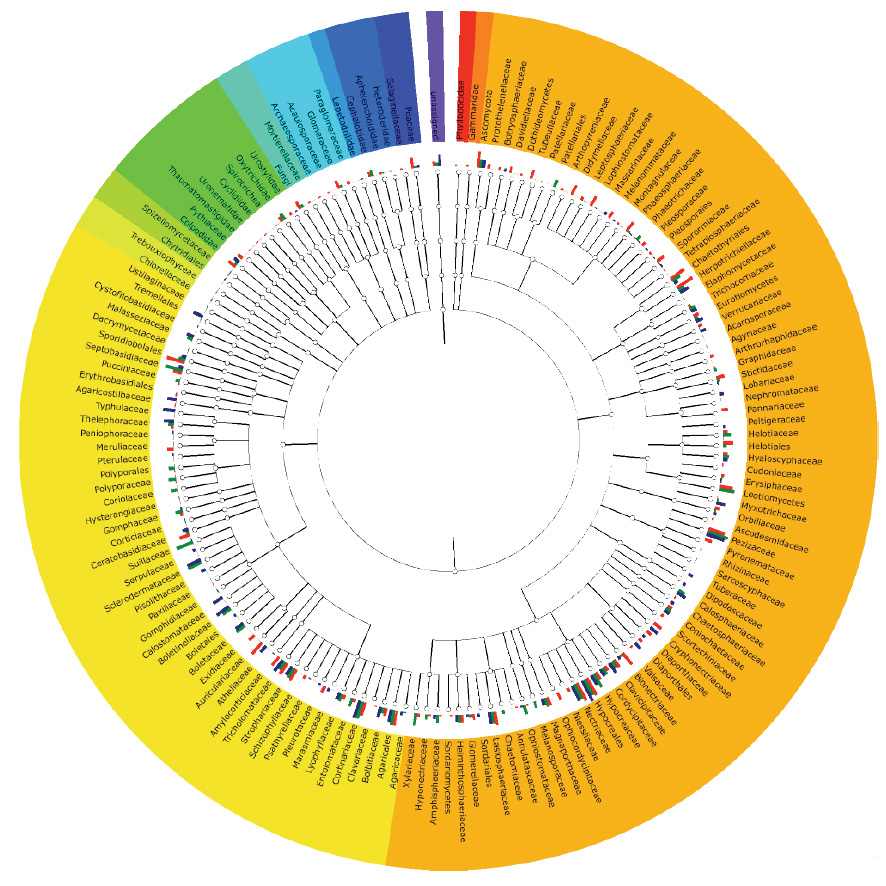
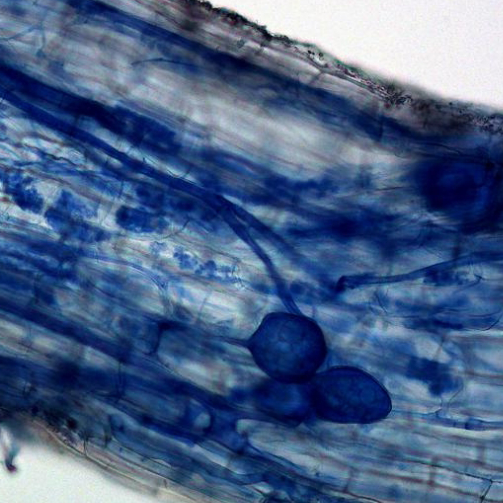
We’re investigating the diversity of both ecomycorrhizal and arbuscular mycorrhizal fungi in relation with host plants.
This diversity is inferred from molecular and morphological characters, as well as community level diversity through amplicon sequencing and longer contigs and scaffolds acquired from metagenome assembly techniques.
We conduct field, greenhouse, and growth chamber studies to understand these symbioses.